3 Introduction to the genetics of taste: PTC
PTC intro adapted from Dolan DNA Learning Center, Cold Spring Harbor Laboratory
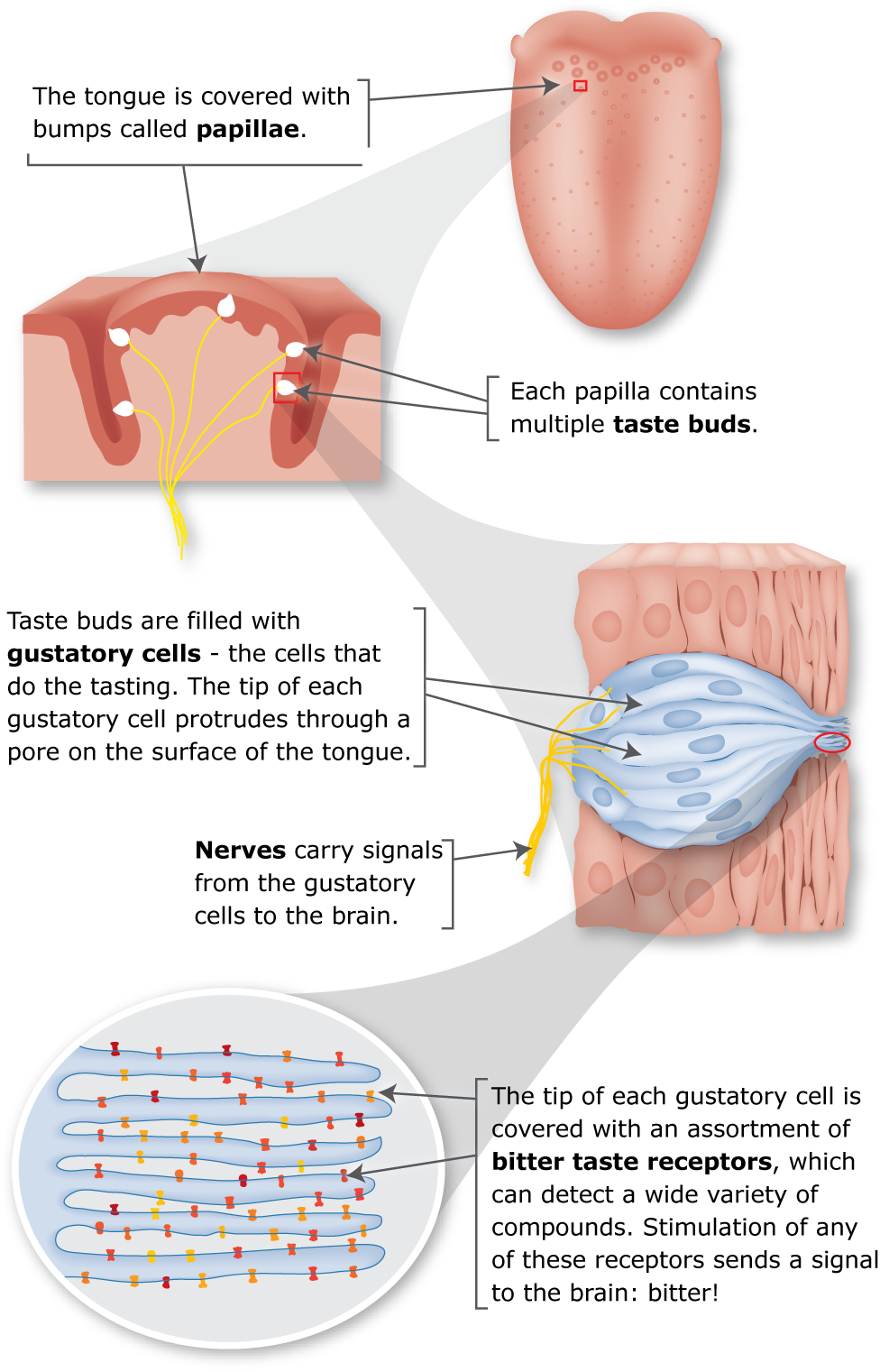
Mammals are believed to distinguish only five basic tastes: sweet, sour, bitter, salty, and umami (the taste of MSG: monosodium glutamate). Taste recognition is mediated by specialized taste cells that communicate with several brain regions through direct connections to sensory neurons. Taste perception is a two-step process. First, a taste molecule binds to a specific receptor on the surface of a taste cell. Then, the taste cell generates a nerve impulse, which is interpreted by the brain. For example, stimulation of “sweet cells” generates a perception of sweetness in the brain. Recent research has shown that taste sensation ultimately is determined by the wiring of a taste cell to the cortex, rather than the type of molecule bound by a receptor. So, for example, if a bitter taste receptor is expressed on the surface of a “sweet cell,” a bitter molecule is perceived as tasting sweet. A serendipitous observation at DuPont, in the early 1930s, first showed a genetic basis to taste. Arthur Fox had synthesized some phenylthiocarbamide (PTC), and some of the PTC dust escaped into the air as he was transferring it into a bottle. Lab-mate C.R. Noller complained that the dust had a bitter taste, but Fox tasted nothing—even when he directly sampled the crystals (Wow..NEVER do this!). Subsequent studies by Albert Blakeslee, at the Carnegie Department of Genetics (the forerunner of Cold Spring Harbor Laboratory), showed that the inability to taste PTC is a recessive trait that varies in the human population.
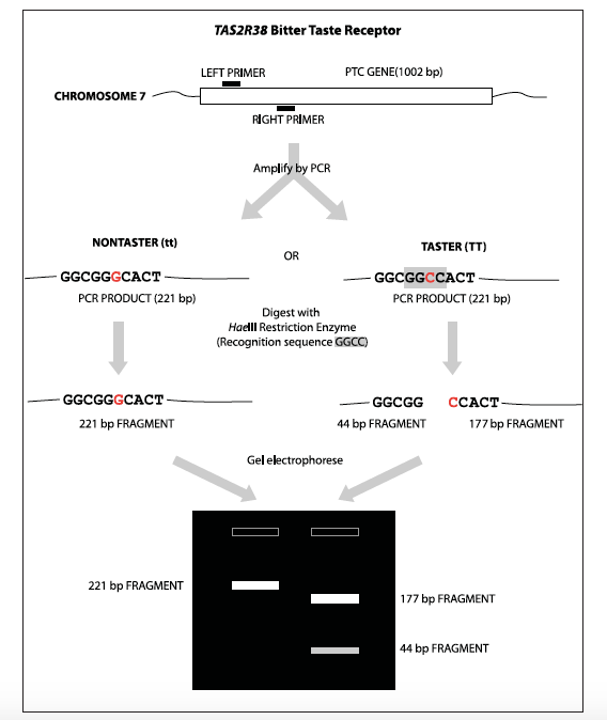
Which gene controls PTC detection?
Bitter-tasting compounds are recognized by receptor proteins on the surface of taste cells. There are approximately 30 genes for different bitter taste receptors in mammals. The gene for the PTC taste receptor, TAS2R38, was identified in 2003. Sequencing identified three nucleotide positions that vary within the human population—each variable position is termed a single nucleotide polymorphism (SNP). One specific combination of the three SNPs, termed a haplotype, correlates most strongly with tasting ability. Analogous changes in other cell-surface molecules influence the activity of many drugs. For example, SNPs in serotonin transporter and receptor genes predict adverse responses to anti-depression drugs, including PROZAC® and Paxil®. In this experiment, a sample of your cheek cells is obtained by saline mouthwash. DNA is extracted from cells via a DNA isolation kit. Polymerase chain reaction (PCR) is then used to amplify a short region of the TAS2R38 gene. The amplified PCR product is digested with the restriction enzyme HaeIII, whose recognition sequence includes one of the SNPs. One allele (a version of the gene) is cut by the enzyme, and one is not—producing a restriction fragment length polymorphism (RFLP) that can be separated on a 2% agarose gel. You will determine your genotype, predict your tasting ability, and taste PTC paper. Class results will show us how well PTC tasting actually conforms to classical Mendelian inheritance, and illustrates the modern concept of pharmacogenetics—where a SNP genotype is used to predict response.
Bitter taste Image from Genetic Science Learning Center. (2016, March 1) PTC The Genetics of Bitter Taste. https://learn.genetics.utah.edu/content/basics/ptc/
