6 PCR purification background
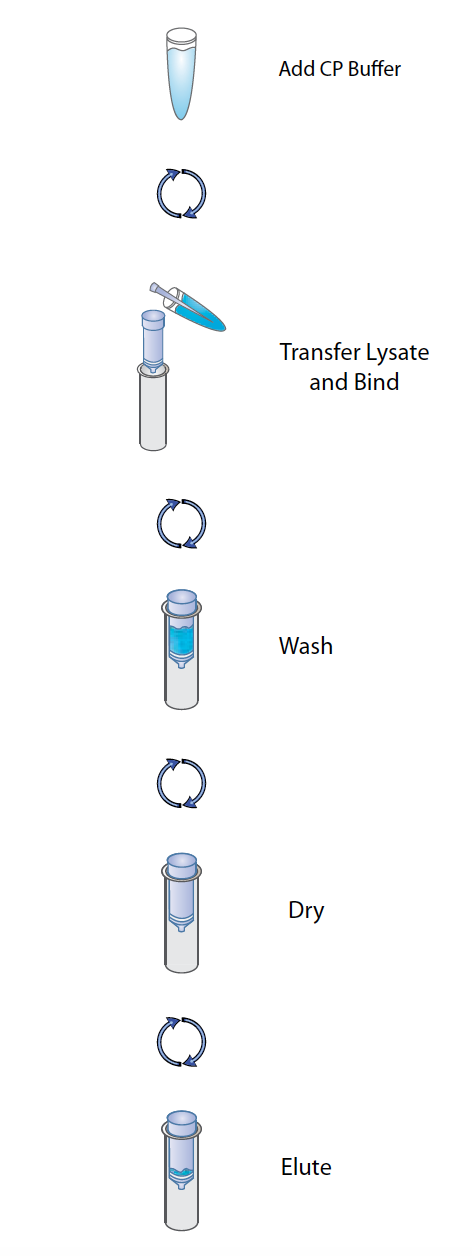
To purify our PCR products, we will bind our DNA to tiny silica columns and perform wash steps and a final elution. You’ll be using a molecular biology ‘kit’–a set of reagents supplied by a company that comes with a simple protocol. These kits are used to increase the efficiency and reproducibility of tasks frequently performed in molecular biology labs. They save you time since all the buffers are supplied for you.
Here is the information from the supplier of this PCR cleanup kit:.
E.Z.N.A.® Cycle Pure Kit simplifies the purification of nucleic acids from a variety of sources. The key to this system is the HiBind® matrix that specifically, but reversibly, binds DNA or RNA under optimized conditions allowing proteins and other contaminants to be removed. Nucleic acids are easily eluted with deionized water or a low salt buffer. The E.Z.N.A.® Cycle Pure Kit is a convenient system for the fast and reliable purification of PCR products. The E.Z.N.A.® Cycle Pure Kit uses HiBind® technology to recover DNA bands from 100 bp to 10 kb free of oligonucleotides, nucleotides, and polymerase with yields exceeding 80%. The binding conditions of the HiBind® DNA Mini Columns are adjusted by the addition of a specially formulated buffer before adding the sample. Following a rapid wash step, DNA is eluted with deionized water or a low salt buffer. Purified DNA can be directly used for most downstream applications include PCR sequencing, restriction enzyme digestion, or various labeling reactions.
I will use 1.5 uL of your sample to determine its concentration on a Nanodrop spectrophotometer.
Why Quantify?
• To check concentration and purity of DNA/RNA present in the solution.
• Determine if samples are useful for downstream applications like: PCR, Restriction digests etc.
Advantages of the nanodrop
• It is a simple machine and economical on space.
• Easy-to-use spectrophotometer
• Can measure small volumes of DNA, RNA and protein concentrations.
NanoDrop Spectrophotometric measures
Measures DNA, RNA (A260) and Proteins (A280) concentrations and sample purity (260:280).
Absorbance at 260 nm
• Nucleic acids absorb UV light at 260 nm due to the aromatic base moieties within their structure. Purines (thymine, cytosine and uracil) and pyrimidines (adenine and guanine) both have peak absorbances at 260 nm, thus making it the standard for quantitating nucleic acid samples.
Absorbance at 280 nm
• The 280 nm absorbance is measured where proteins and phenolic compounds have a strong absorbance. Similarly, the aromaticity of phenol groups of organic compounds absorbs strongly near 280 nm.
Absorbance at 230 nm
• Many organic compounds have strong absorbances at around 225 nm. In addition to phenol, TRIzol, and chaotropic salts, the peptide bonds in proteins absorb light between 200 and 230 nm.
NanoDrop Spectrophometric measurements
• The A260/280 ratio shows the purity of the sample analyzed.
|
Nucleic Acid Type
|
Approximate A260/A280 Ratio of pure sample
|
|
Pure DNA
|
1.8
|
|
Pure RNA
|
2.0
|
|
Pure Protein
|
0.57
|
• A260/230 ratio indicates the presence of organic contaminants, such as (but not limited to): phenol, TRIzol, chaotropic salts and other aromatic compounds.
• Samples with 260/230 ratios below 1.8 are considered to have a significant amount of these contaminants
Note that: Because of the low concentrations, sometimes it is difficult to assess the purity of the samples (esp. RNA) by analyzing the A260/280 and A260/230 ratios. Quantification of very low concentration samples cannot be assessed by the NanoDrop because they are outside the lowest concentrations the NanoDrop is designed to measure. A more sensitive method such as an Agilent Bioanalyzer or Qubit analysis is recommended.

